Alpha SC - Efficient GPU-accelerated single-cell data analysis
Alpha SC, the most efficient GPU-accelerated single-cell data analysis pipeline from BioTuring Alpha, is an innovative initiative by BioTuring designed to address the challenges of analyzing large-scale biological data. Alpha SC is expected to boost the efficiency of single-cell data analysis, laying the foundation for a revolutionary shift for scientists to analyze large single-cell datasets in realtime.
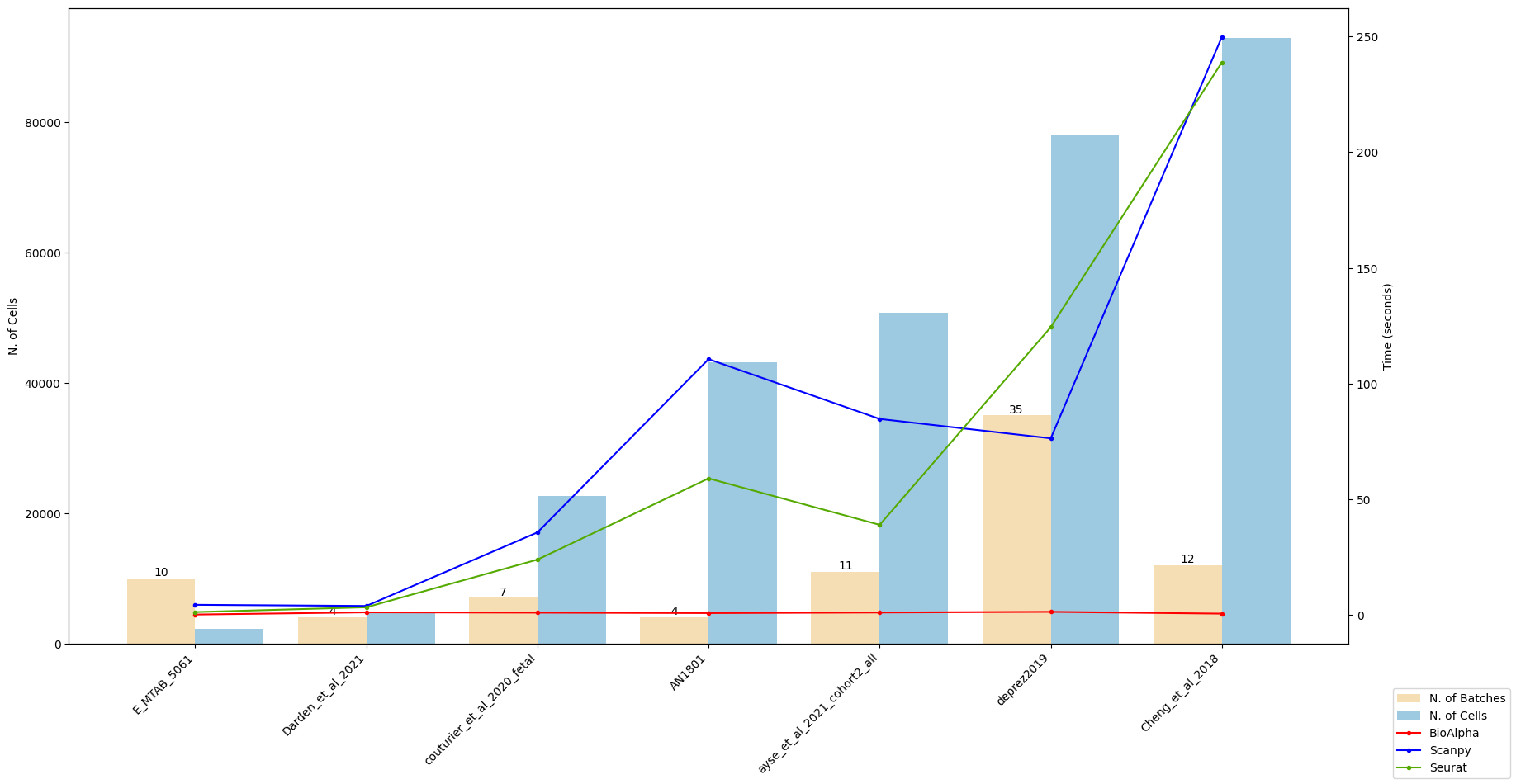
Impressive benchmark results
For the same pipeline, BioTuring Alpha’s single-cell pipeline has reported an end-to-end runtime that was 169 times and 121 times faster than Scanpy and Seurat, respectively.
AUCell
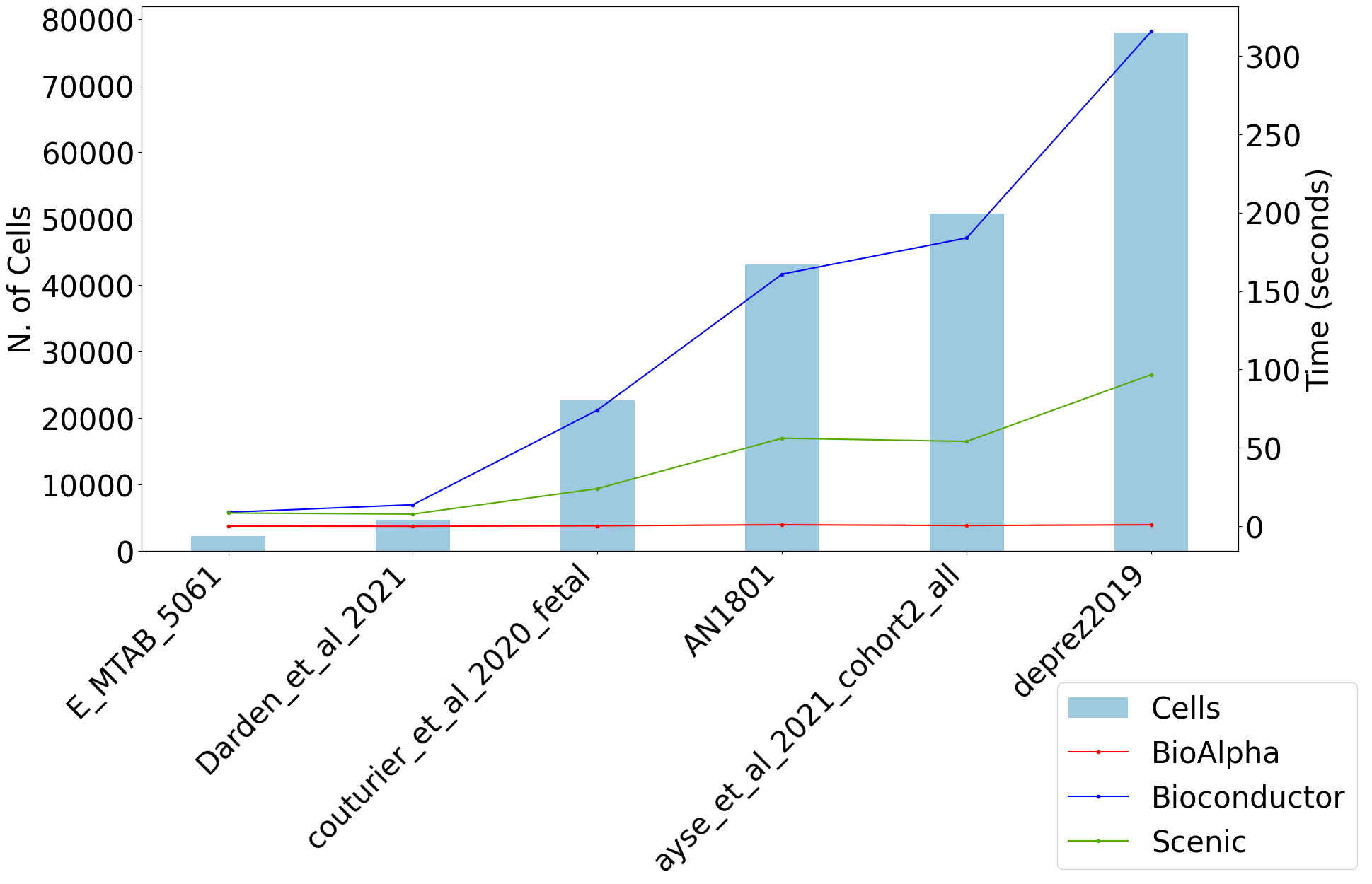
t-SNE
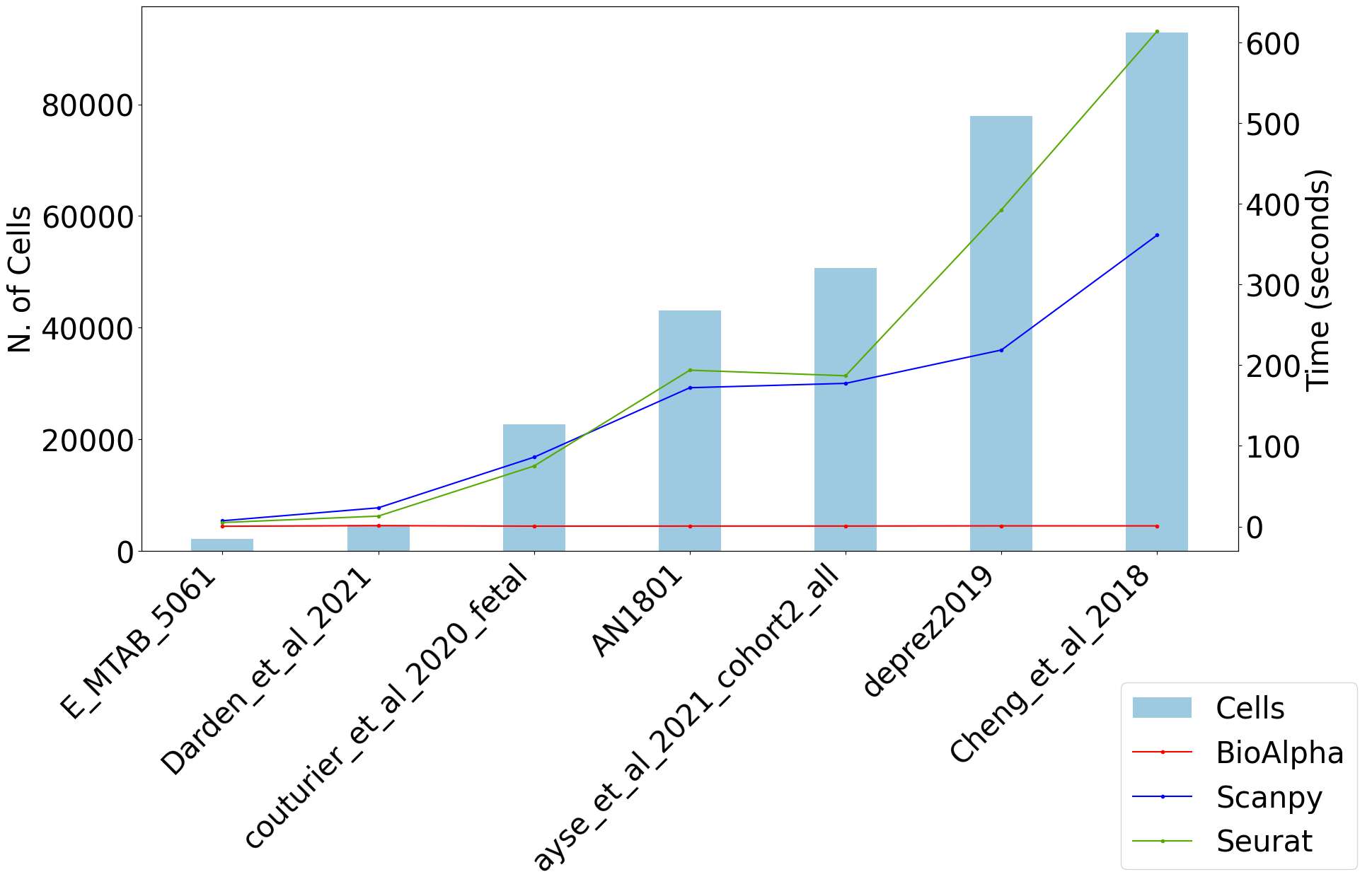
UMAP
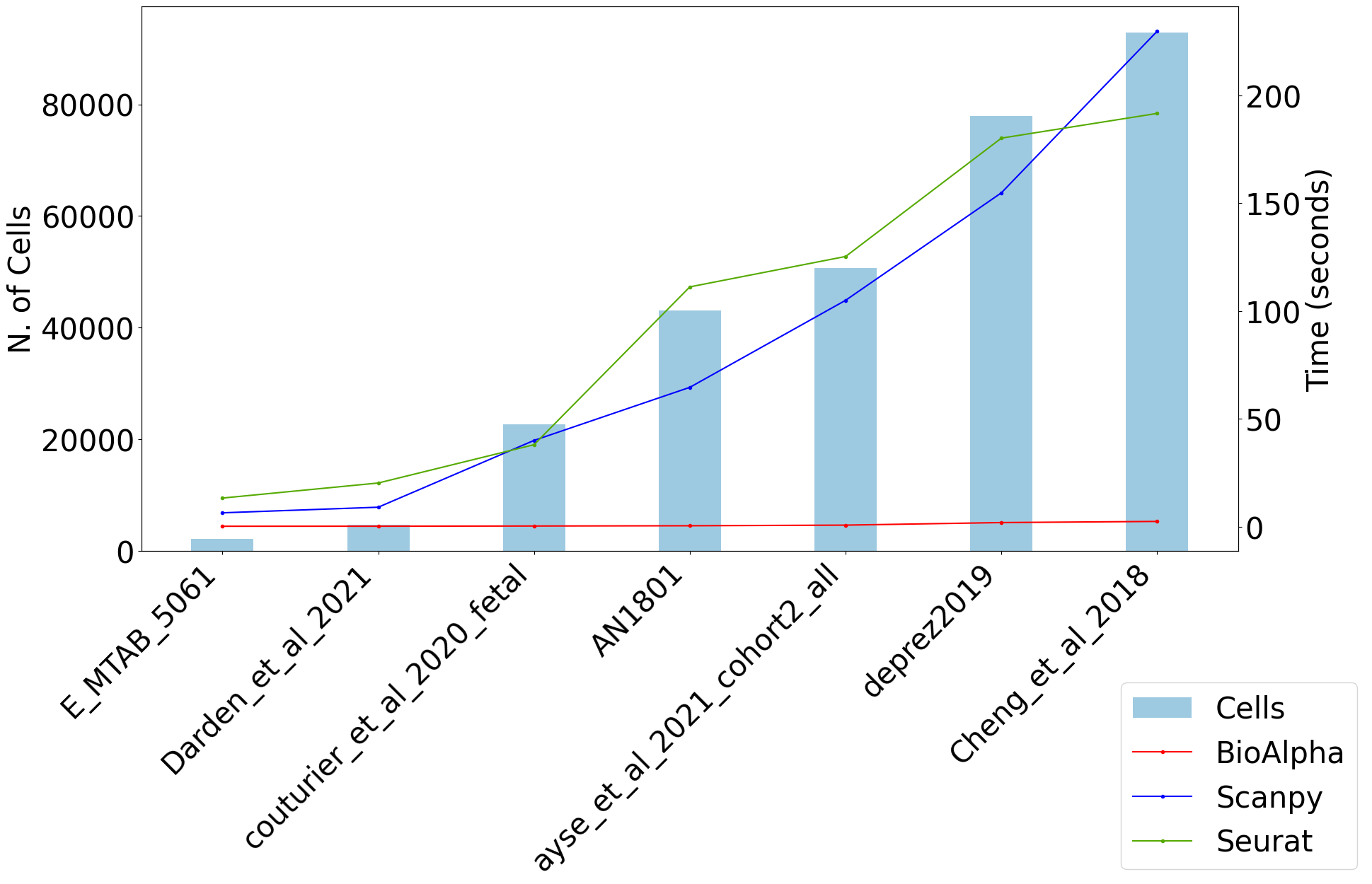
Louvain
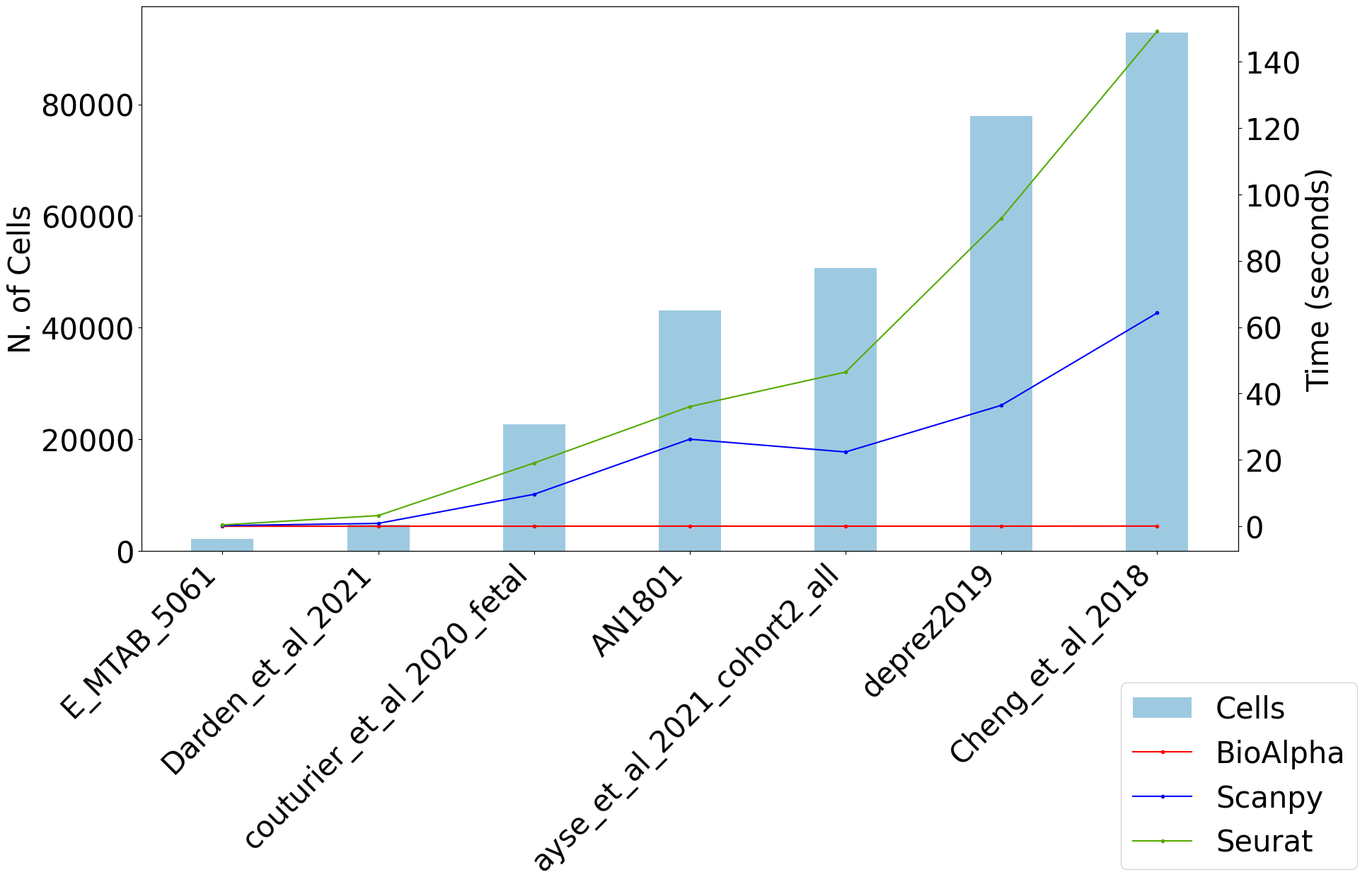
Harmony
Features
Read and Write a Single-Cell Matrix
Principal Component Analysis (PCA)
Geometric sketching
Harmony
k-Nearest neighbors (k-NN)
Louvain Alpha
t-SNE (t-distributed Stochastic Neighbor Embedding)
UMAP (Uniform Manifold Approximation and Projection)
Venice
AUCell enrichment analysis
Getting started
Academic users
We offer a version that is free for academic researchers. Don’t hesitate to download the package and start experimenting with your data. If you wish to upgrade to the professional version, please contact us at support@bioturing.com.
Enterprise
Our pipeline can be seamlessly integrated into your company’s internal framework, providing valuable assistance to your bioinformatics team. We offer extensive support to ensure smooth operations. For more information, please contact us at support@bioturing.com.
Usage
Get started by browsing Installation, Tutorial, or the main API.